Data analysis
Download the script for this session at the link below (Click Link –> Right Click on Page –> Save Page As):
Basic data analysis
R can be used for basic arithmetic and it can also be used as a calculator. We start the session with doing basic computations.
1+3## [1] 41+(4*2)## [1] 91+(4^2) ##^: exponent## [1] 17Trigonometric functions
sin(30)## [1] -0.9880316log10 (10)## [1] 1log10 (20)## [1] 1.30103Comparisons
2 == 2## [1] TRUE2 == 4## [1] FALSE1 < 2## [1] TRUEData analysis using a dataframe
R can also be used for more complex computations which involve statistical tests. These are used to analyse our data after cleaning and visualising our data. In this section, we will continue to work with the penguins data from the data visualisation section. These data have already been cleaned so we will go straight into analysis. load the processed data
#### Extra packages
install.packages('broom')
install.packages('stargazer')
install.packages('effects')
#### Load libraries
library(readxl)
library(tidyverse)
library(janitor)
library(dplyr)
library(palmerpenguins)
library(broom)
library(stargazer)
library(effects)
library(ggplot2)#### Load data ----
penguins <- palmerpenguins::penguins
str(penguins)## tibble [344 × 8] (S3: tbl_df/tbl/data.frame)
## $ species : Factor w/ 3 levels "Adelie","Chinstrap",..: 1 1 1 1 1 1 1 1 1 1 ...
## $ island : Factor w/ 3 levels "Biscoe","Dream",..: 3 3 3 3 3 3 3 3 3 3 ...
## $ bill_length_mm : num [1:344] 39.1 39.5 40.3 NA 36.7 39.3 38.9 39.2 34.1 42 ...
## $ bill_depth_mm : num [1:344] 18.7 17.4 18 NA 19.3 20.6 17.8 19.6 18.1 20.2 ...
## $ flipper_length_mm: int [1:344] 181 186 195 NA 193 190 181 195 193 190 ...
## $ body_mass_g : int [1:344] 3750 3800 3250 NA 3450 3650 3625 4675 3475 4250 ...
## $ sex : Factor w/ 2 levels "female","male": 2 1 1 NA 1 2 1 2 NA NA ...
## $ year : int [1:344] 2007 2007 2007 2007 2007 2007 2007 2007 2007 2007 ...View(penguins)#### Selecting and filtering data
penguins_bill <- select (penguins, species, island, bill_length_mm, bill_depth_mm)
penguins_flipper <- select (penguins, species, island, flipper_length_mm)
penguins_bodymass <- select(penguins, -flipper_length_mm, -bill_length_mm, -bill_depth_mm)
penguins_09 <- filter(penguins, year == 2009)
penguins2 <- filter(penguins, year < 2008)
#### say for instance you want the penguin bill information for 2007 and 2008 only
##### option 1
penguins_bill2 <- select (penguins, species, island, bill_length_mm, bill_depth_mm, year)
penguins_bill2 <- filter(penguins_bill2, year < 2009)
penguins_bill2## # A tibble: 224 × 5
## species island bill_length_mm bill_depth_mm year
## <fct> <fct> <dbl> <dbl> <int>
## 1 Adelie Torgersen 39.1 18.7 2007
## 2 Adelie Torgersen 39.5 17.4 2007
## 3 Adelie Torgersen 40.3 18 2007
## 4 Adelie Torgersen NA NA 2007
## 5 Adelie Torgersen 36.7 19.3 2007
## 6 Adelie Torgersen 39.3 20.6 2007
## 7 Adelie Torgersen 38.9 17.8 2007
## 8 Adelie Torgersen 39.2 19.6 2007
## 9 Adelie Torgersen 34.1 18.1 2007
## 10 Adelie Torgersen 42 20.2 2007
## # … with 214 more rowspenguins_bill2 <- na.omit(penguins_bill2)
penguins_bill2## # A tibble: 223 × 5
## species island bill_length_mm bill_depth_mm year
## <fct> <fct> <dbl> <dbl> <int>
## 1 Adelie Torgersen 39.1 18.7 2007
## 2 Adelie Torgersen 39.5 17.4 2007
## 3 Adelie Torgersen 40.3 18 2007
## 4 Adelie Torgersen 36.7 19.3 2007
## 5 Adelie Torgersen 39.3 20.6 2007
## 6 Adelie Torgersen 38.9 17.8 2007
## 7 Adelie Torgersen 39.2 19.6 2007
## 8 Adelie Torgersen 34.1 18.1 2007
## 9 Adelie Torgersen 42 20.2 2007
## 10 Adelie Torgersen 37.8 17.1 2007
## # … with 213 more rows##### option 2
penguins_bill2_2 <- select(filter(penguins, year < 2009), species, island, bill_length_mm, bill_depth_mm, year)
penguins_bill2_2 <- na.omit(penguins_bill2_2)
penguins_bill2_2## # A tibble: 223 × 5
## species island bill_length_mm bill_depth_mm year
## <fct> <fct> <dbl> <dbl> <int>
## 1 Adelie Torgersen 39.1 18.7 2007
## 2 Adelie Torgersen 39.5 17.4 2007
## 3 Adelie Torgersen 40.3 18 2007
## 4 Adelie Torgersen 36.7 19.3 2007
## 5 Adelie Torgersen 39.3 20.6 2007
## 6 Adelie Torgersen 38.9 17.8 2007
## 7 Adelie Torgersen 39.2 19.6 2007
## 8 Adelie Torgersen 34.1 18.1 2007
## 9 Adelie Torgersen 42 20.2 2007
## 10 Adelie Torgersen 37.8 17.1 2007
## # … with 213 more rows##### option 3
## you can use the pipe function
Penguins_bill<-penguins %>%
filter(year < 2009) %>%
select(species, island, bill_length_mm, bill_depth_mm, year) %>%
na.omit()
Penguins_bill## # A tibble: 223 × 5
## species island bill_length_mm bill_depth_mm year
## <fct> <fct> <dbl> <dbl> <int>
## 1 Adelie Torgersen 39.1 18.7 2007
## 2 Adelie Torgersen 39.5 17.4 2007
## 3 Adelie Torgersen 40.3 18 2007
## 4 Adelie Torgersen 36.7 19.3 2007
## 5 Adelie Torgersen 39.3 20.6 2007
## 6 Adelie Torgersen 38.9 17.8 2007
## 7 Adelie Torgersen 39.2 19.6 2007
## 8 Adelie Torgersen 34.1 18.1 2007
## 9 Adelie Torgersen 42 20.2 2007
## 10 Adelie Torgersen 37.8 17.1 2007
## # … with 213 more rows##### mutate
Penguins_bill<-Penguins_bill %>% mutate(bill_length_m = bill_length_mm/1000) #added a new column with new units for bill_length
Penguins_bill## # A tibble: 223 × 6
## species island bill_length_mm bill_depth_mm year bill_length_m
## <fct> <fct> <dbl> <dbl> <int> <dbl>
## 1 Adelie Torgersen 39.1 18.7 2007 0.0391
## 2 Adelie Torgersen 39.5 17.4 2007 0.0395
## 3 Adelie Torgersen 40.3 18 2007 0.0403
## 4 Adelie Torgersen 36.7 19.3 2007 0.0367
## 5 Adelie Torgersen 39.3 20.6 2007 0.0393
## 6 Adelie Torgersen 38.9 17.8 2007 0.0389
## 7 Adelie Torgersen 39.2 19.6 2007 0.0392
## 8 Adelie Torgersen 34.1 18.1 2007 0.0341
## 9 Adelie Torgersen 42 20.2 2007 0.042
## 10 Adelie Torgersen 37.8 17.1 2007 0.0378
## # … with 213 more rowsPenguins_bill<-Penguins_bill %>% mutate(bill_depth_m = bill_depth_mm/1000)
##### group_by
Penguins_bill_mean<-Penguins_bill %>%
group_by(species, year) %>%
summarise(mean_bill_length_mm = mean(bill_length_mm))
Penguins_bill_mean## # A tibble: 6 × 3
## # Groups: species [3]
## species year mean_bill_length_mm
## <fct> <int> <dbl>
## 1 Adelie 2007 38.8
## 2 Adelie 2008 38.6
## 3 Chinstrap 2007 48.7
## 4 Chinstrap 2008 48.7
## 5 Gentoo 2007 47.0
## 6 Gentoo 2008 46.9Penguins_bill_mean<-Penguins_bill %>%
group_by(species, year) %>%
summarise(mean_bill_length_mm = mean(bill_length_mm)) %>%
arrange(desc(year))
Penguins_bill_mean## # A tibble: 6 × 3
## # Groups: species [3]
## species year mean_bill_length_mm
## <fct> <int> <dbl>
## 1 Adelie 2008 38.6
## 2 Chinstrap 2008 48.7
## 3 Gentoo 2008 46.9
## 4 Adelie 2007 38.8
## 5 Chinstrap 2007 48.7
## 6 Gentoo 2007 47.0Penguins_bill_mean<-Penguins_bill %>%
group_by(species, year) %>%
summarise(mean_bill_length_mm = mean(bill_length_mm))
Penguins_bill_mean## # A tibble: 6 × 3
## # Groups: species [3]
## species year mean_bill_length_mm
## <fct> <int> <dbl>
## 1 Adelie 2007 38.8
## 2 Adelie 2008 38.6
## 3 Chinstrap 2007 48.7
## 4 Chinstrap 2008 48.7
## 5 Gentoo 2007 47.0
## 6 Gentoo 2008 46.9alpha=0.05
Penguins_bill_Stats <- Penguins_bill %>%
group_by(species, island, year) %>%
summarise(
n=n(),
mean_bill_length_mm=mean(na.omit(bill_length_mm)),
bill_length_sd=sd(na.omit(bill_length_mm)),
mean_bill_depth_mm=mean(na.omit(bill_length_mm)),
bill_depth_sd=sd(na.omit(bill_length_mm)))
Penguins_bill_Stats## # A tibble: 10 × 8
## # Groups: species, island [5]
## species island year n mean_bill_lengt… bill_length_sd mean_bill_depth…
## <fct> <fct> <int> <int> <dbl> <dbl> <dbl>
## 1 Adelie Biscoe 2007 10 38.3 1.85 38.3
## 2 Adelie Biscoe 2008 18 38.7 2.59 38.7
## 3 Adelie Dream 2007 20 39.1 2.32 39.1
## 4 Adelie Dream 2008 16 38.2 2.79 38.2
## 5 Adelie Torge… 2007 19 38.8 2.93 38.8
## 6 Adelie Torge… 2008 16 38.8 3.65 38.8
## 7 Chinstrap Dream 2007 26 48.7 3.47 48.7
## 8 Chinstrap Dream 2008 18 48.7 3.62 48.7
## 9 Gentoo Biscoe 2007 34 47.0 3.27 47.0
## 10 Gentoo Biscoe 2008 46 46.9 2.64 46.9
## # … with 1 more variable: bill_depth_sd <dbl>##### Plot stats
names(Penguins_bill_Stats)## [1] "species" "island" "year"
## [4] "n" "mean_bill_length_mm" "bill_length_sd"
## [7] "mean_bill_depth_mm" "bill_depth_sd"StatsPlot<-ggplot(Penguins_bill_Stats, aes(x = species, y = mean_bill_length_mm))+
geom_line(size = 1, colour = "red")+
geom_point(shape = 19, size = 1, colour = "red")+
geom_errorbar(aes(ymin=mean_bill_length_mm-bill_length_sd, ymax=mean_bill_length_mm+bill_length_sd),width=.5, colour ="black", linetype = "solid")
StatsPlot<-ggplot(Penguins_bill_Stats, aes(x = species, y = mean_bill_length_mm))+
geom_line(size = 1, colour = "red")+
geom_point(shape = 19, size = 1, colour = "red")+
geom_errorbar(aes(ymin=mean_bill_length_mm-bill_length_sd, ymax=mean_bill_length_mm+bill_length_sd),width=.5, colour ="black", linetype = "solid")+
theme_classic()+
xlab("Species_Names")+
ylab("Bill_length (mm)")
StatsPlot<-ggplot(Penguins_bill_Stats, aes(x = species, y = mean_bill_length_mm))+
geom_line(size = 1, colour = "red")+
geom_point(shape = 19, size = 1, colour = "red")+
geom_errorbar(aes(ymin=mean_bill_length_mm-bill_length_sd, ymax=mean_bill_length_mm+bill_length_sd),width=.5, colour ="black", linetype = "solid")+
theme_classic()+
xlab("Species_Names")+
ylab("Bill_length (mm)")+facet_grid(rows=vars(year))
StatsPlot<-ggplot(Penguins_bill_Stats, aes(x = species, y = mean_bill_length_mm))+
geom_line(size = 8, colour = "red")+
geom_point(shape = 19, size = 4, colour = "red")+
geom_errorbar(aes(ymin=mean_bill_length_mm-bill_length_sd, ymax=mean_bill_length_mm+bill_length_sd),width=.1, colour ="black", linetype = "solid")+
theme_classic()+
xlab("Species names")+
ylab("Bill length (mm)")+facet_grid(rows=vars(year, island))
StatsPlot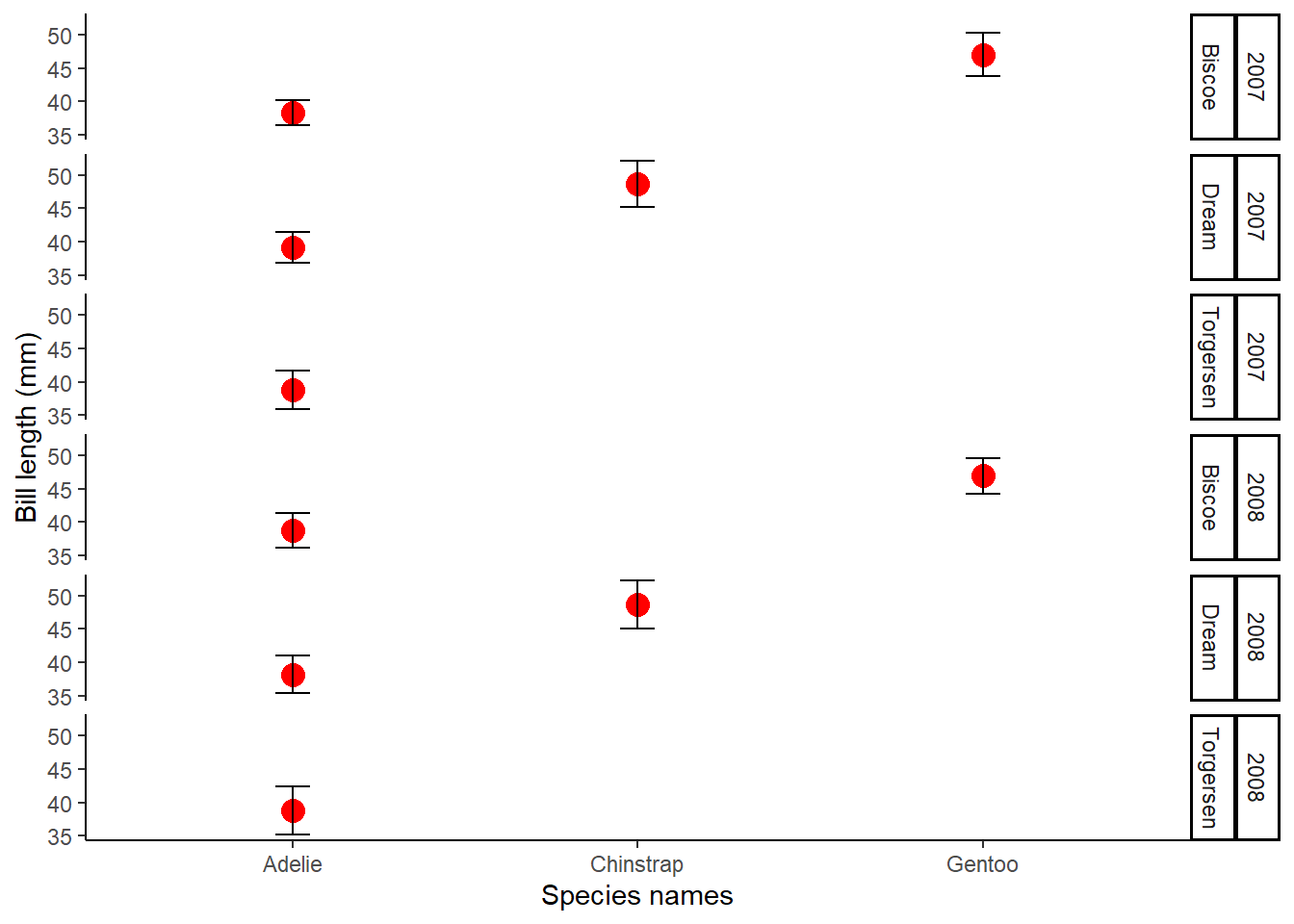
StatsPlot2<-ggplot(Penguins_bill_Stats, aes(x = species, y = mean_bill_length_mm, col=island))+
geom_line(size = 8)+
geom_point(shape = 19, size = 4)+
geom_errorbar(aes(ymin=mean_bill_length_mm-bill_length_sd, ymax=mean_bill_length_mm+bill_length_sd),width=.1, colour ="black", linetype = "solid")+
theme_classic()+
xlab("Species names")+
ylab("Bill length (mm)")+facet_grid(rows=vars(year))
StatsPlot2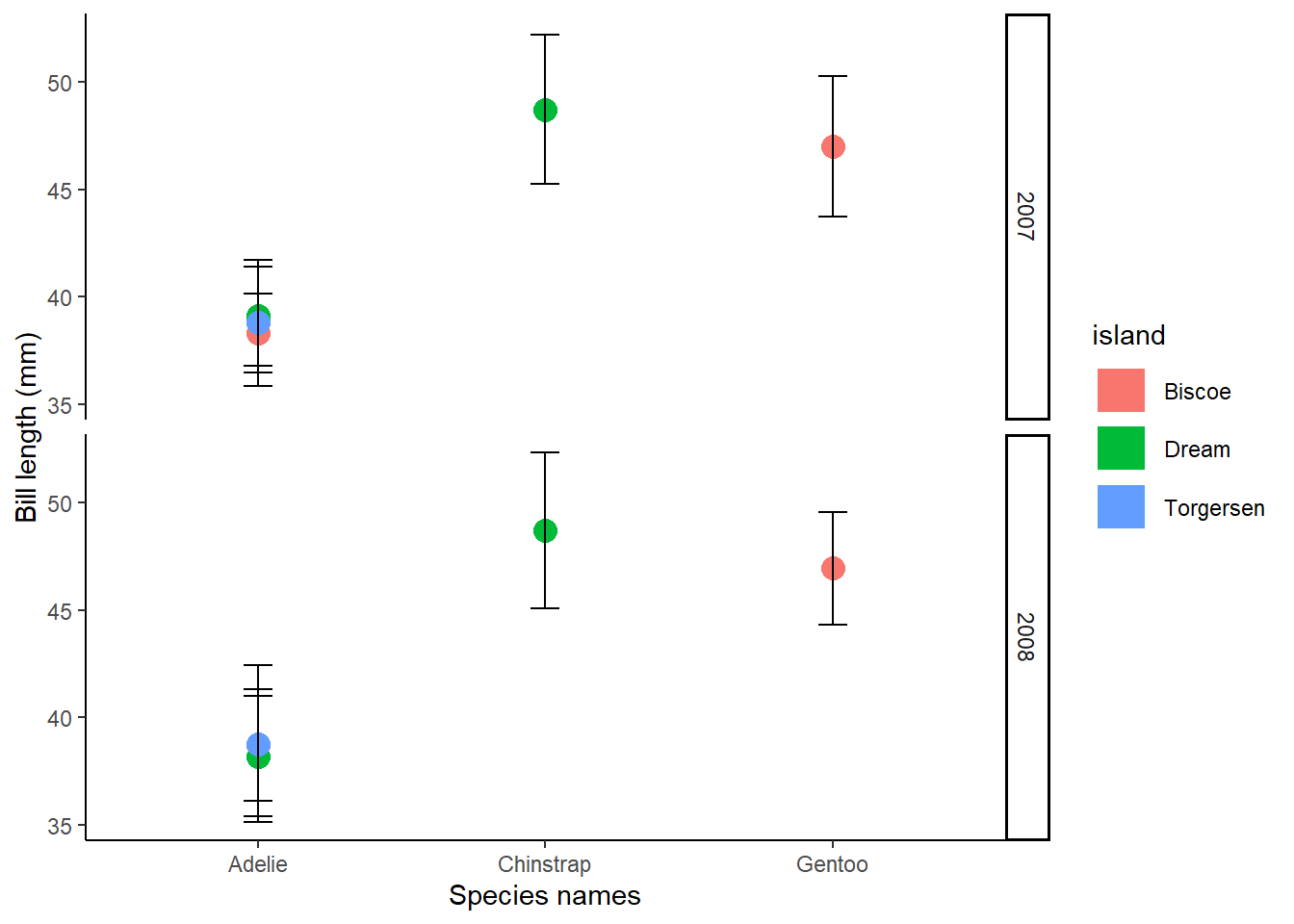
#### Categorical linear model ----
lm <- lm(bill_length_mm ~ island, data = penguins)
summary(lm)##
## Call:
## lm(formula = bill_length_mm ~ island, data = penguins)
##
## Residuals:
## Min 1Q Median 3Q Max
## -12.0677 -3.8559 0.2958 3.8175 14.3425
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 45.2575 0.3897 116.127 < 2e-16 ***
## islandDream -1.0897 0.5970 -1.825 0.0688 .
## islandTorgersen -6.3065 0.8057 -7.827 6.44e-14 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 5.036 on 339 degrees of freedom
## (2 observations deleted due to missingness)
## Multiple R-squared: 0.154, Adjusted R-squared: 0.149
## F-statistic: 30.86 on 2 and 339 DF, p-value: 4.86e-13#### Analysis of Variance ----
aov <- aov(bill_length_mm ~ island, data = penguins)
aov## Call:
## aov(formula = bill_length_mm ~ island, data = penguins)
##
## Terms:
## island Residuals
## Sum of Squares 1565.599 8598.607
## Deg. of Freedom 2 339
##
## Residual standard error: 5.03633
## Estimated effects may be unbalanced
## 2 observations deleted due to missingnesssummary(aov)## Df Sum Sq Mean Sq F value Pr(>F)
## island 2 1566 782.8 30.86 4.86e-13 ***
## Residuals 339 8599 25.4
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
## 2 observations deleted due to missingness##### Post-hoc test - if there is a significant difference
Tukey.aov <- TukeyHSD(aov)
Tukey.aov## Tukey multiple comparisons of means
## 95% family-wise confidence level
##
## Fit: aov(formula = bill_length_mm ~ island, data = penguins)
##
## $island
## diff lwr upr p adj
## Dream-Biscoe -1.089743 -2.495170 0.3156833 0.1628734
## Torgersen-Biscoe -6.306505 -8.203279 -4.4097305 0.0000000
## Torgersen-Dream -5.216762 -7.188974 -3.2445487 0.0000000#### Continuous linear model ----
lm2 <- lm(bill_length_mm ~ body_mass_g, data = penguins)
summary(lm2)##
## Call:
## lm(formula = bill_length_mm ~ body_mass_g, data = penguins)
##
## Residuals:
## Min 1Q Median 3Q Max
## -10.1251 -3.0434 -0.8089 2.0711 16.1109
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 2.690e+01 1.269e+00 21.19 <2e-16 ***
## body_mass_g 4.051e-03 2.967e-04 13.65 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 4.394 on 340 degrees of freedom
## (2 observations deleted due to missingness)
## Multiple R-squared: 0.3542, Adjusted R-squared: 0.3523
## F-statistic: 186.4 on 1 and 340 DF, p-value: < 2.2e-16#### Categorical and Continuous model ----
lm3 <- lm(bill_length_mm ~ body_mass_g + island, data = penguins)
summary(lm3)##
## Call:
## lm(formula = bill_length_mm ~ body_mass_g + island, data = penguins)
##
## Residuals:
## Min 1Q Median 3Q Max
## -10.4676 -2.3806 0.0498 2.4179 13.8966
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 21.7428965 1.6468756 13.203 < 2e-16 ***
## body_mass_g 0.0049861 0.0003431 14.532 < 2e-16 ***
## islandDream 3.9118982 0.5818015 6.724 7.56e-11 ***
## islandTorgersen -1.2723006 0.7216507 -1.763 0.0788 .
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 3.957 on 338 degrees of freedom
## (2 observations deleted due to missingness)
## Multiple R-squared: 0.4793, Adjusted R-squared: 0.4747
## F-statistic: 103.7 on 3 and 338 DF, p-value: < 2.2e-16#### Summarise results ----
lm_coefficients <- tidy(lm)
lm2_coefficients <- tidy(lm2)
lm3_coefficients <- tidy(lm3)#### Plot effects ----
lm_effects <- allEffects(lm)
lm2_effects <- allEffects(lm2)
lm3_effects <- allEffects(lm3)
aov_effects <- allEffects(aov)
plot(lm_effects)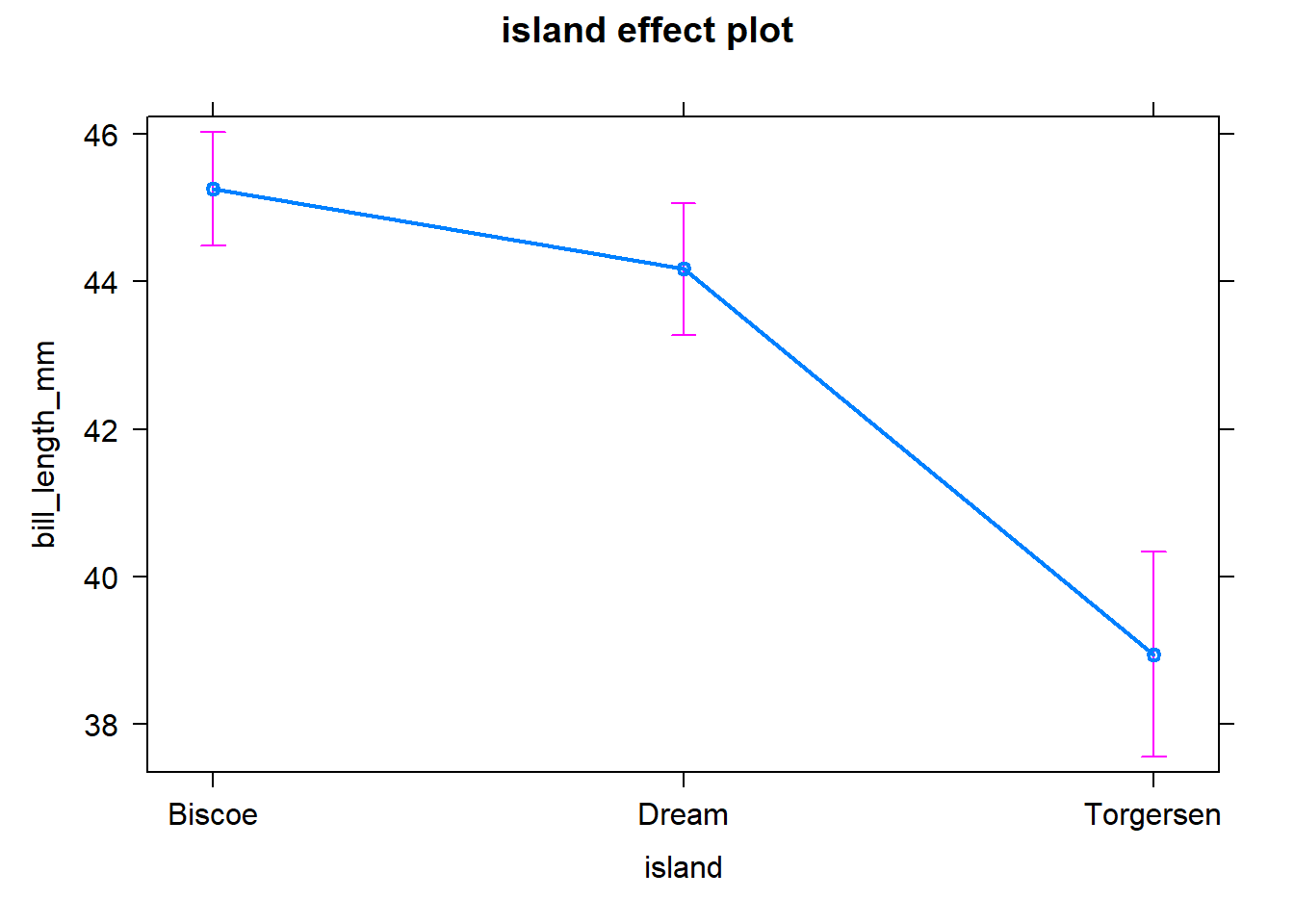
plot(lm2_effects)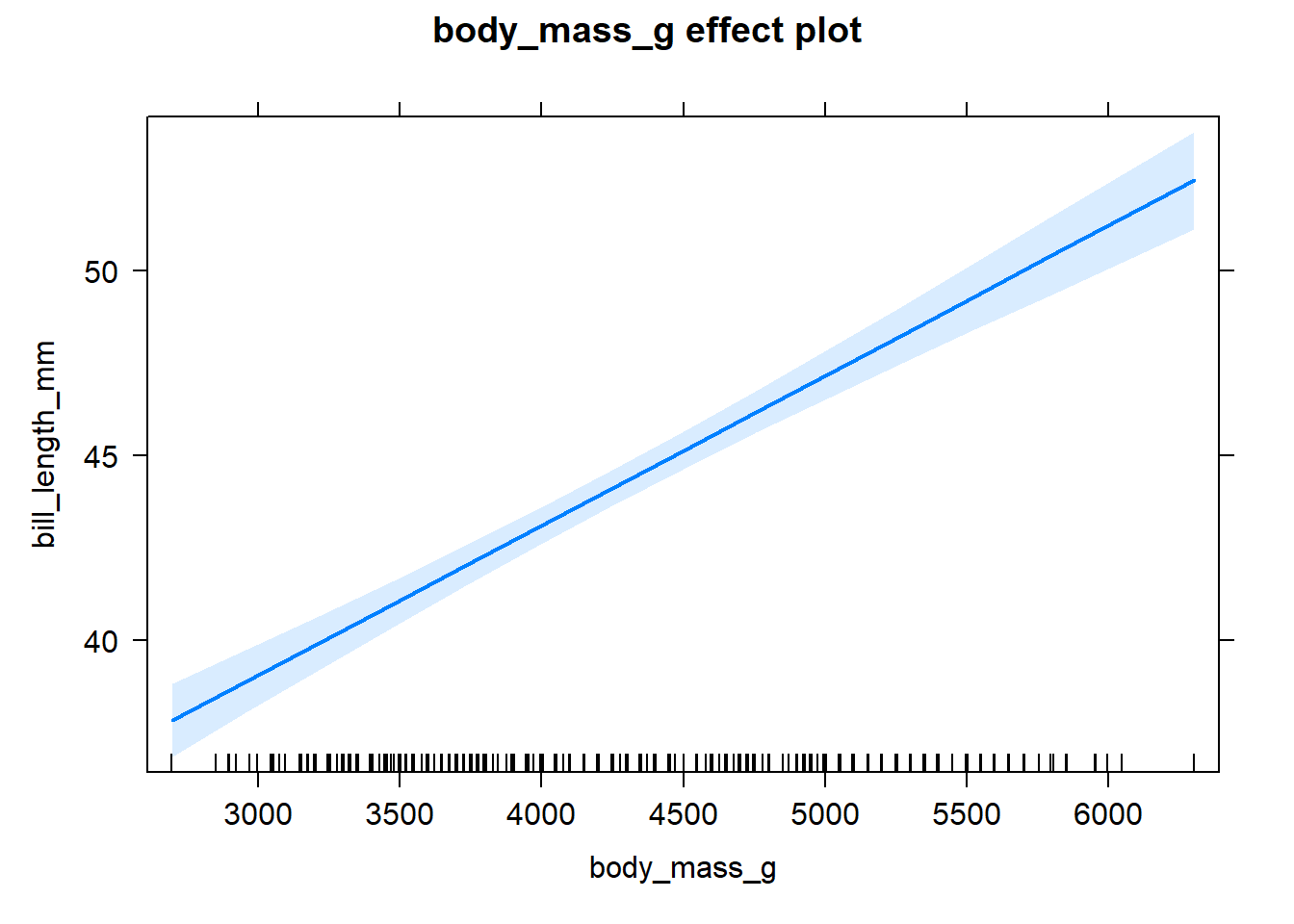
plot(lm3_effects)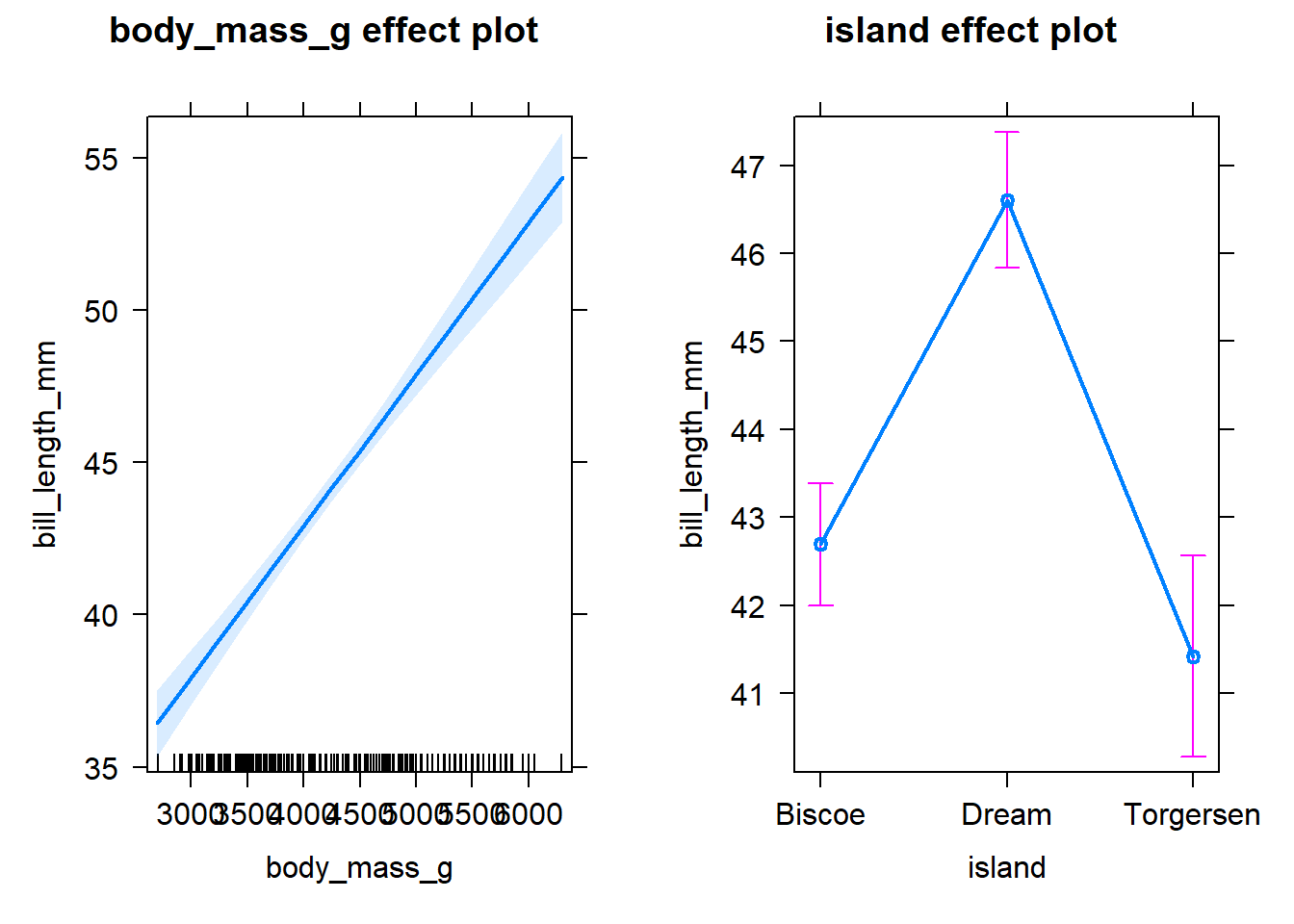
plot(aov_effects)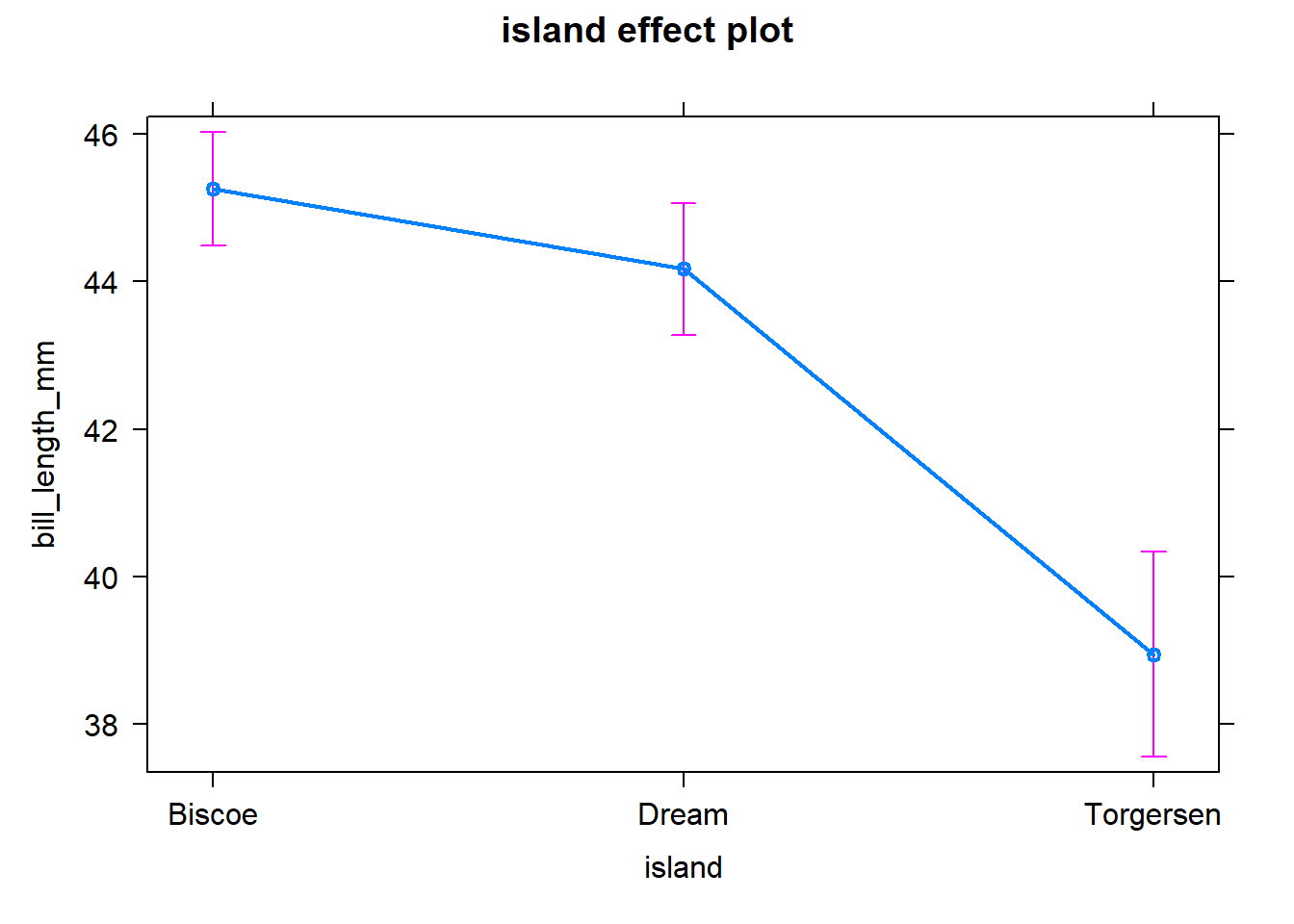
#### Stargazer summary table ----
# export as a text; copy and paste
stargazer::stargazer(lm, lm2, lm3, type = 'text')### you can do this for the aov too##
## =============================================================================================
## Dependent variable:
## -------------------------------------------------------------------------
## bill_length_mm
## (1) (2) (3)
## ---------------------------------------------------------------------------------------------
## islandDream -1.090* 3.912***
## (0.597) (0.582)
##
## islandTorgersen -6.307*** -1.272*
## (0.806) (0.722)
##
## body_mass_g 0.004*** 0.005***
## (0.0003) (0.0003)
##
## Constant 45.257*** 26.899*** 21.743***
## (0.390) (1.269) (1.647)
##
## ---------------------------------------------------------------------------------------------
## Observations 342 342 342
## R2 0.154 0.354 0.479
## Adjusted R2 0.149 0.352 0.475
## Residual Std. Error 5.036 (df = 339) 4.394 (df = 340) 3.957 (df = 338)
## F Statistic 30.862*** (df = 2; 339) 186.443*** (df = 1; 340) 103.720*** (df = 3; 338)
## =============================================================================================
## Note: *p<0.1; **p<0.05; ***p<0.01